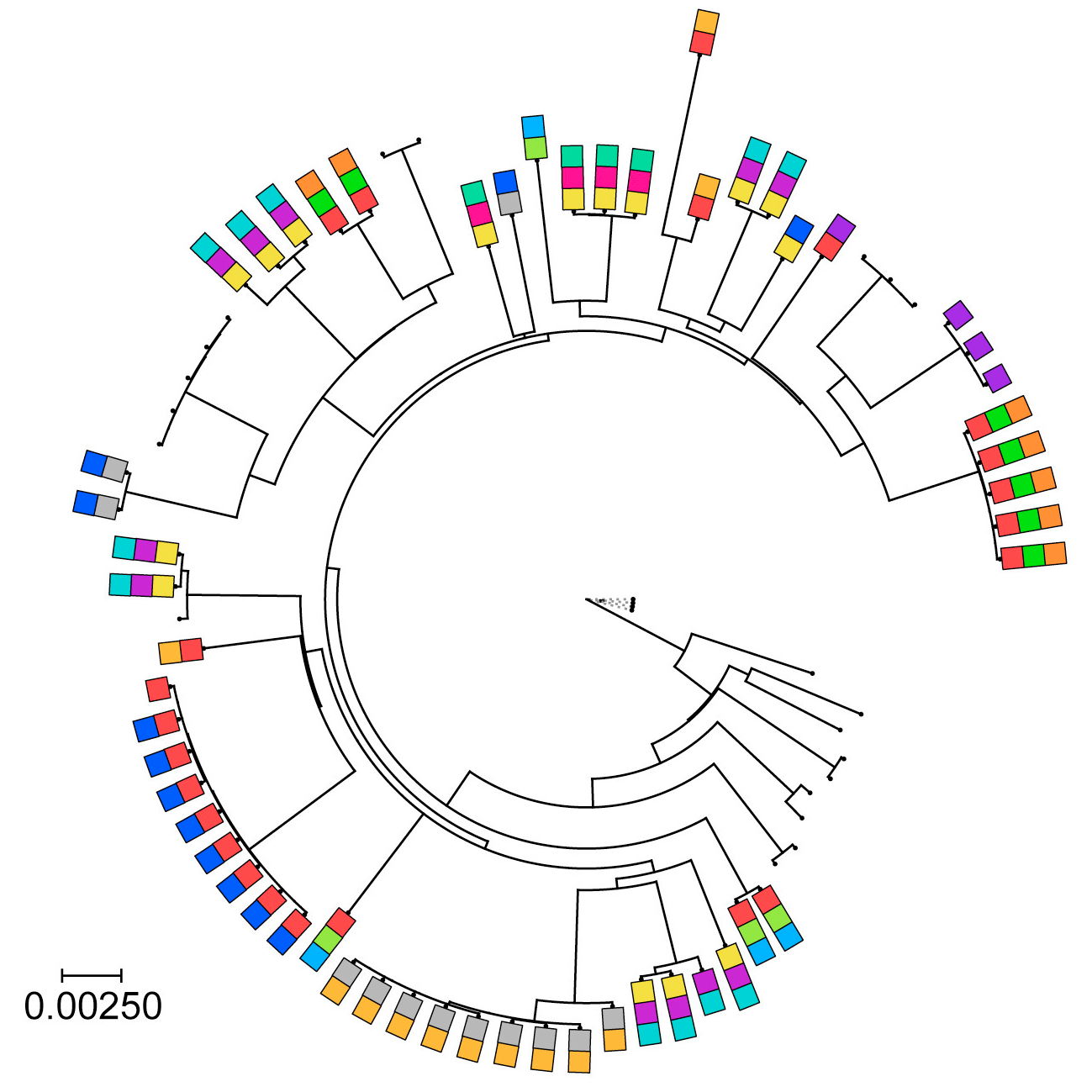
A phylogenetic tree of B. fragilis with colored boxes that indicate the presence of different type 6 secretion system toxins. [Pubmed]

A phylogenetic tree of B. fragilis with colored boxes that indicate the presence of different type 6 secretion system toxins. [Pubmed]
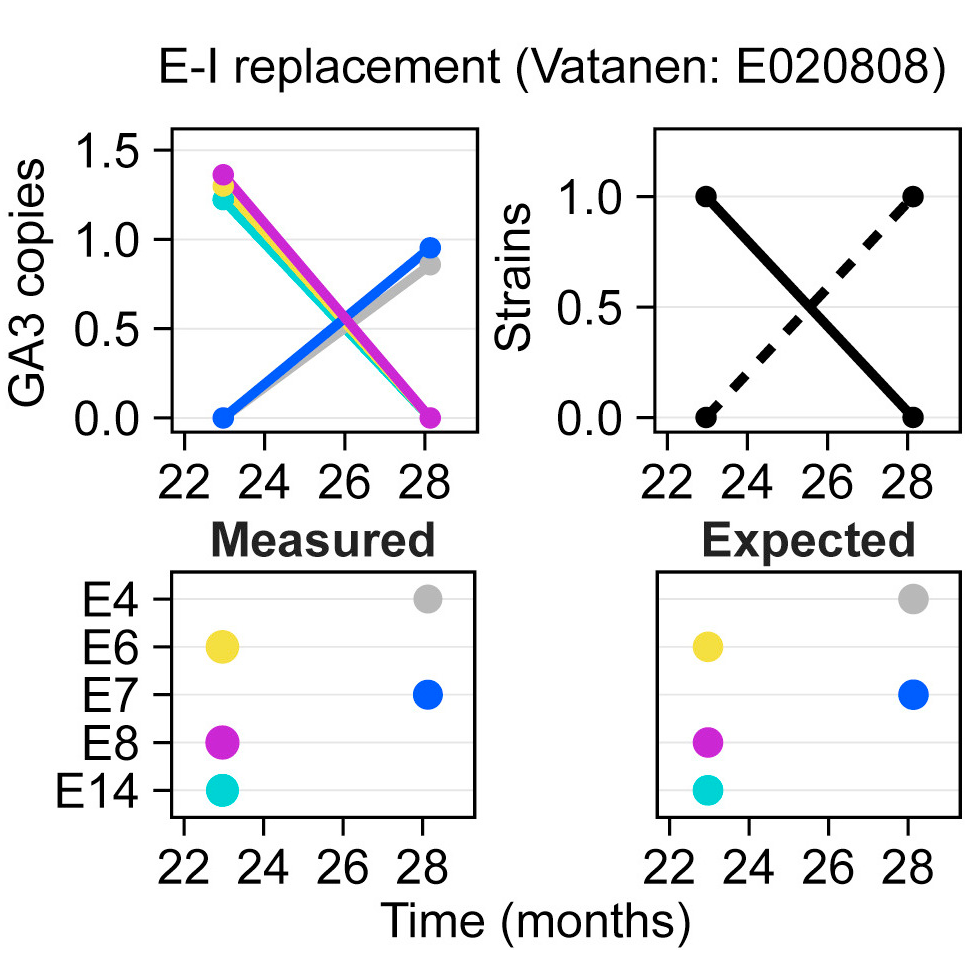
These graphs show a change in the B. fragilis strain during microbiome development. A change in the toxin gene matches a change in the inferred strain. [Pubmed]
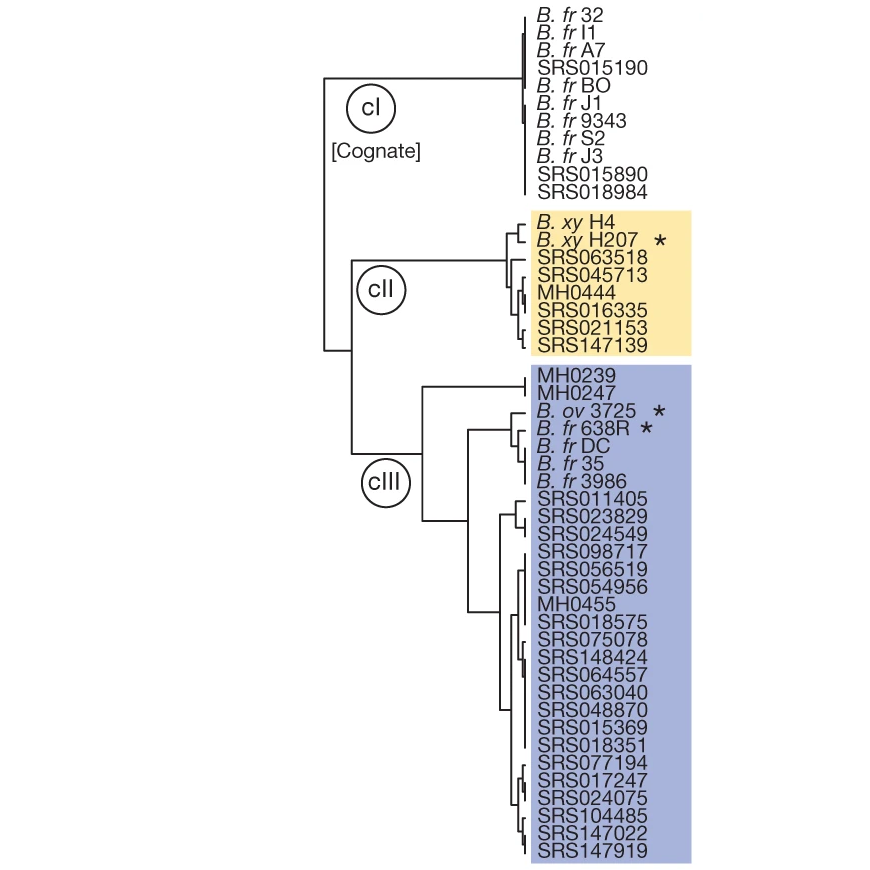
A phylogenetic tree of a type 6 secretion system immunity gene from sequences genomes as well as from public metagenomes (starting with SRS or MH). [Pubmed]
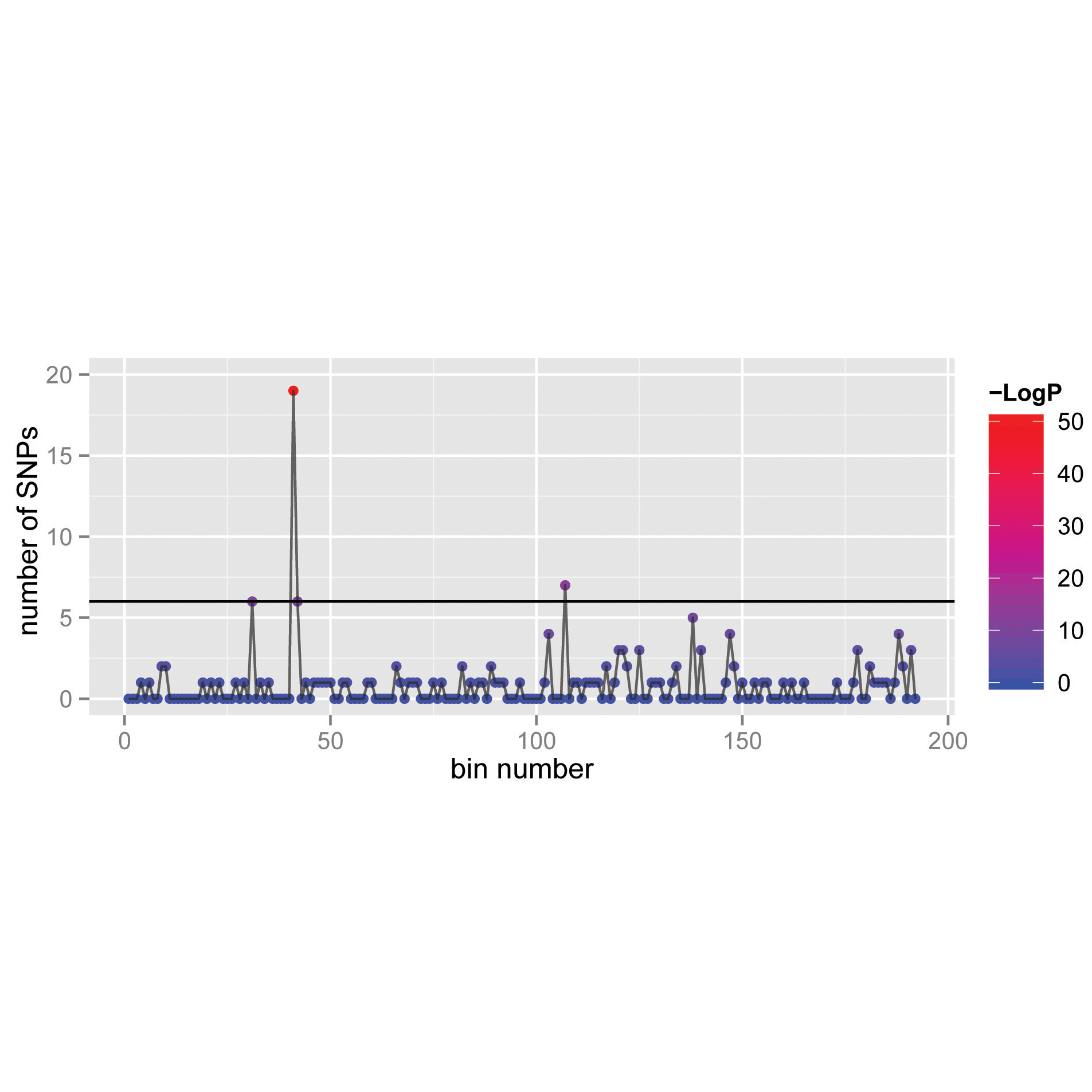
The number of nucleotide variants identified along the sequence a viral genome. Each bin is associated with a p-value that highlights regions with a concentration of variants. [Pubmed]
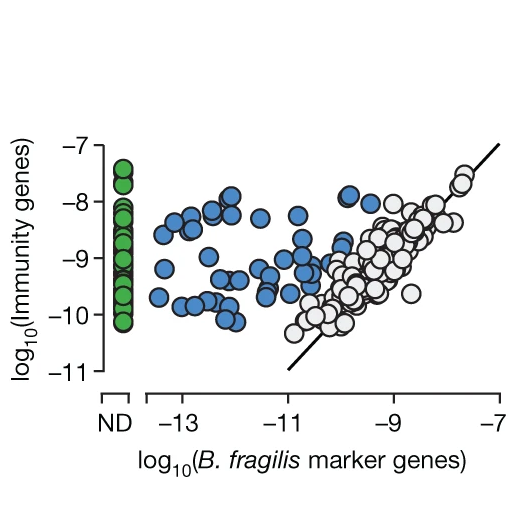
The relationship between B. fragilis levels and immune gene levels in the microbiome. There are three populations, first there are cases when B. fragilis and the immune genes are on par (grey) which is expected, second there are cases where the immune genes are much higher than the species (blue), third there are cases where the immune gene is found and B. fragilis is absent (green). [Pubmed]
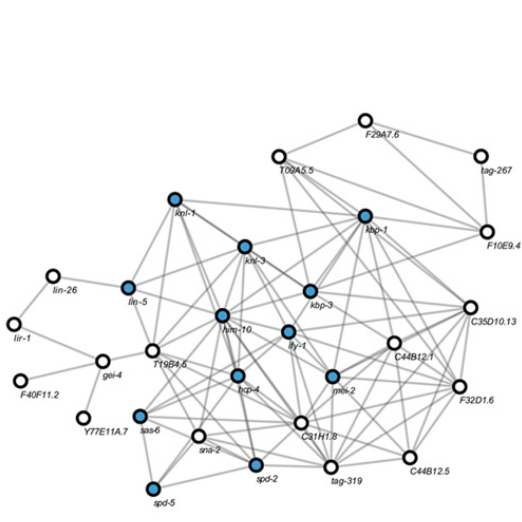
A network that shows the relationships between novel genes in C. elegans. The network is Wormnet which has been derived from multiple sources of genomic information. Nodes colored in blue are associated with the centromere. [Pubmed]
Verster A.J, Petronella N., Green J., Matias F., Brooks S.P.J. (2022). A Bayesian method for identifying associations between response variables and bacterial community composition. PLoS Computational Biology. 18(7):e1010108. [Pubmed]
Ng V., Rees E.E., Niu J. Zaghool, A., Ghiasbeglou H., Verster A. (2020). Application of natural language processing algorithms for extracting information from news articles in event-based surveillance. Canada Communicable Disease Report. 46(6):186-191. [Pubmed]
Eng A., Verster A.J., Borenstein E. (2020). MetaLAFFA: a flexible, end-to-end, distributed computing-compatible metagenomic functional annotation pipeline. BMC Bioinformatics. 21(1):471. [Pubmed]
Pilar A.V.C., Petronella N., Dussault F.M., Verster A.J., Bekal S., Levesque R.C., Goodridge L., Tamber S. (2020). Similar yet different: phylogenomic analysis to delineate Salmonella and Citrobacter species boundaries. BMC Genomics. 21(1):377. [Pubmed]
Ross B.D.*, Verster A.J.*, Radey M.C., Schmidtke D.T., Pope C.E., Hoffman L.R., Hajjar A.M., Peterson S.B., Borenstein E., Mougous J.D. (2019). Human gut bacteria contain acquired interbacterial defence systems. Nature. 575(7781):224-228. [Pubmed]
Verster A.J., Borenstein E. (2018). Competitive lottery-based assembly of selected clades in the human gut microbiome. Microbiome. 6(1):186. [Pubmed]
Verster A.J.*, Ross B.D.*, Radey M.C., Bao Y., Goodman A., Mougous J.D., Borenstein E. (2017). The Landscape of Type VI Secretion across Human Gut Microbiomes Reveals its Role in Community Composition. Cell Host and Microbe. 22(3):411-419. [Pubmed]
Verster A.J., Styles E.B., Mateo A., Derry B.W, Andrews B.F., Fraser A.G. (2017). Taxonomically restricted genes with essential functions play a role in chromosome segregation in C. elegans and S. cerevisiae. Genes | Genomes | Genetics. 7(10):3337-3347. [Pubmed]
Whitney J.C., Peterson S.B., Kim J., Pazos M., Verster A.J., Radey M.C., Kulasekara H.D., Ching M.Q., Bullen N.P., Bryant D., Goo Y.A., Surette M.G., Borenstein E., Vollmer W., Mougous J.D. (2017). A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. ELife. 6:e26938. [Pubmed]
Styles E.B., Founk K. J., Zamparo L.A., Sing T.L., Altintas D., Ribeyre C., Ribaud V., Rougemont J., Mayhew D., Costanzo M., Usaj M., Verster A.J., Koch E.N., Novarina D., Graf M., Luke B., Muzi-Falconi M., Myers C.L., Mitra R.D., Shore D., Brown G.W., Zhang Z., Boone C., Andrews B.J. (2016). Exploring Quantitative Yeast Phenomics with Single-Cell Analysis of DNA Damage Foci. Cell Systems. 3(3):264-277. [Pubmed]
Usaj M., Styles E.B., Verster A.J., Friesen H., Boone C., Andrews B.J. (2016). High-Content Screening for Quantitative Cell Biology. Trends in Cell Biology. 26(8):598-611 [Pubmed]
Salonen J.S., Verster A.J., Engels E., Soininen J., Luoto M. (2016). Calibrating aquatic microfossil proxies with regression-tree ensembles: cross-validation with modern chironomid and diatom data. The Holocene. 26(7):1040-1048
Vu V., Verster A.J., Schertzberg M., Chuluunbaatar T., Spensley M., Pakic D., Hart T., Moffat J., Fraser A.G. (2015). RNAi screens in C. elegans reveal that natural variation in gene expression predicts the severity of mutant phenotypes. Cell. 162(2):391-402 [Pubmed]
Smithson C., Purdy A., Verster A.J., Upton C. (2014). Prediction of Steps in the Evolution of Variola Virus Host Range. PLoS One. 9(3): e91520. [Pubmed]
Verster A.J., Ramani A.K., McKay S.J., Fraser A.G. (2014). Comparative RNAi screens in C. elegans and C. briggsae reveal the impact of Developmental System Drift on gene function. PLoS Genetics. 10(2):e1004077 [Pubmed]
Ramani A.K., Chuluunbaatar T., Verster A.J., Na H., Vu V., Pelte N., Wannissorn N., Jiao A., Fraser A.G. (2012). The majority of animal genes are required for wild-type fitness. Cell. 148(4):792-802 [Pubmed]